Genomics of Comparative Seed Evolution
The Genomics of Comparative Seed Evolution
Stemming from an original grant awarded in 2004 by the NSF, this grant from 2011 has been an ongoing project to exploit plant genome diversity to discover new genes involved in the development of seeds. This project has generated new data, resources, and bioinformatic tools and analytical pipelines to enable functional trait-to-gene predictions for any species based on phylogeny and/or machine learning approaches. It is envisioned that the data and software resources generated will empower Comparative Genomic researchers to exploit plant diversity to identify genes associated with any trait of interest or economic value. The current portion of the project involves the RNA sequencing of the most primitive living seed plants, the non-flowering Gymnosperms, on earth since the Mesozoic era. This research transcends and unites genomics and systematics to produce a deeper understanding of seed evolution, while revealing genes involved in important agronomic traits involved in seed development. The findings will have implications for improving seed quality, as much of today’s agriculture – from food to textiles- depends on seed products.
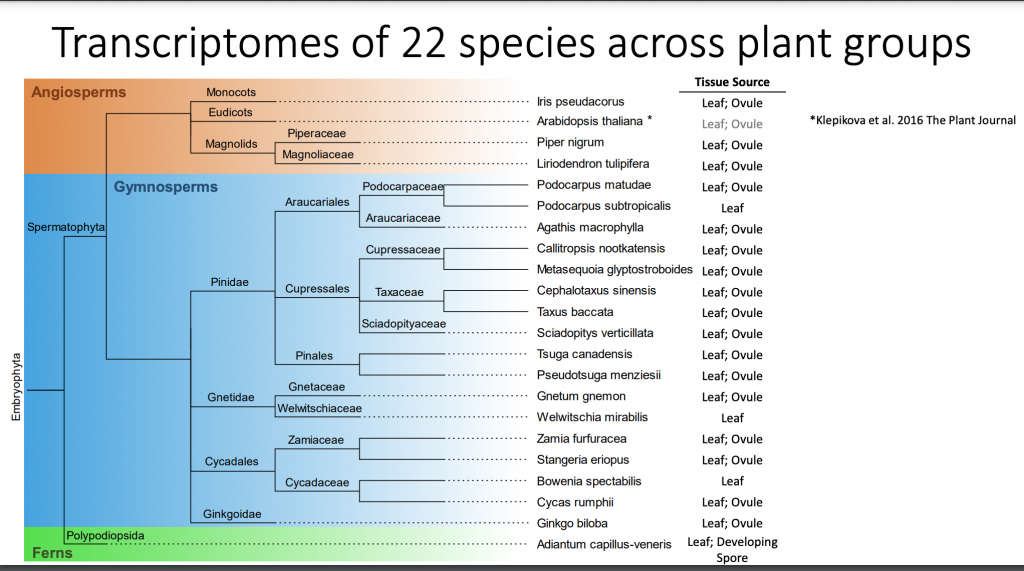
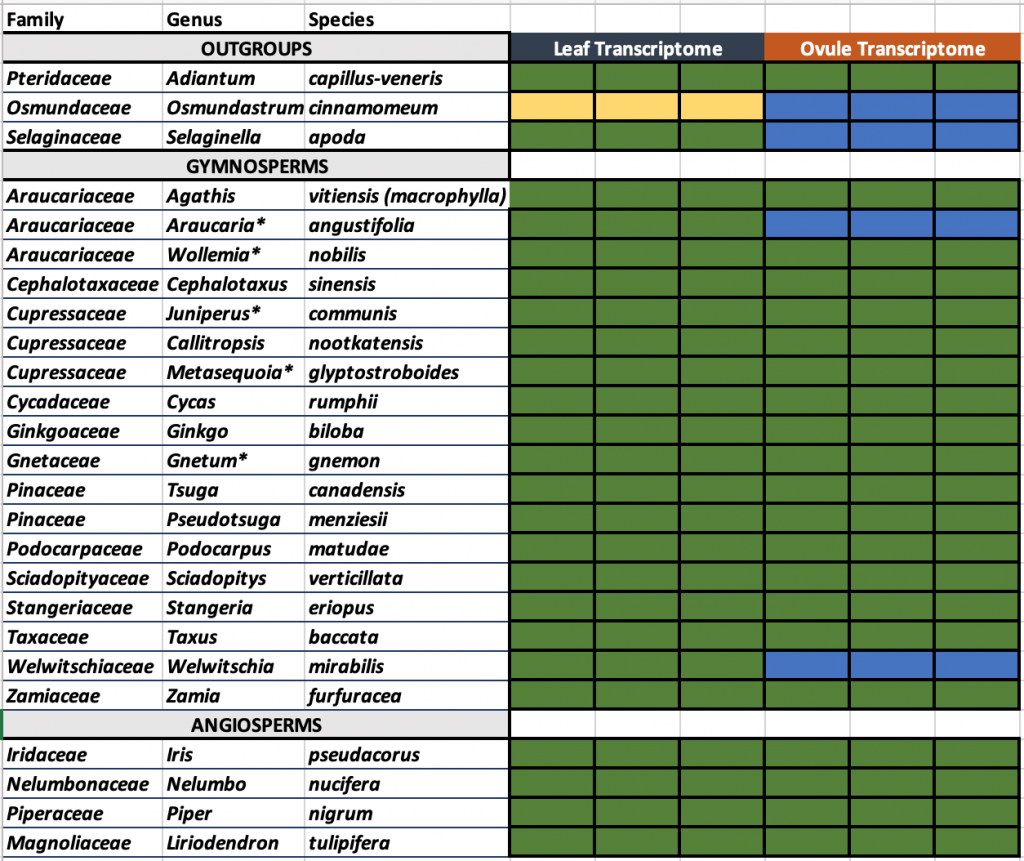
This project includes the deep RNA sequencing of 18 Gymnosperm species, as well as four Angiosperms and three ferns species, which serve as outgroups.

Functional Genomics of Seed Evolution Data Links
- BigPlant website with alignment and trees
- Gikgo biloba cDNA Illumina reads (SRX022254 & SRX022196)
- de la Torre-Bárcena et al. (2009, PLoS ONE): amino acid alignment; ML tree (Newick format, PDF graphic)
- de la Torre et al. (2006, BMC Evolutionary Biology): amino acid alignment; MP tree (Newick format, PDF graphic)
- EST data
For More Information about the seed project, please contact Dennis Stevenson – DWS@NYBG.ORG